Predict protein domain families from coding transcripts
predictDomains.RdPredict protein domain families from coding transcripts
predictDomains(x, fasta, ..., plot = FALSE, progress_bar = FALSE, ncores = 4)Arguments
- x
Can be a GRanges object containing 'CDS' features in GTF format
Can be a GRangesList object containing CDS ranges for each transcript
- fasta
BSgenome or Biostrings object containing genomic sequence
- ...
Logical conditions to pass to dplyr::filter to subset transcripts for analysis. Variables are metadata information found in `x` and multiple conditions can be provided delimited by comma. Example: transcript_id == "transcript1"
- plot
Argument whether to plot out protein domains (Default: FALSE). Note: only first 20 proteins will be plotted
- progress_bar
Argument whether to show progress bar (Default: FALSE). Useful to track progress of predicting a long list of proteins.
- ncores
Number of cores to utilise to perform prediction
Value
Dataframe containing protein features for each cds entry
Examples
## ---------------------------------------------------------------------
## EXAMPLE USING SAMPLE DATASET
## ---------------------------------------------------------------------
# Load Mouse genome sequence
library(BSgenome.Mmusculus.UCSC.mm10)
# Load dataset
data(new_query_gtf)
# predict domains of all CDSs in query GTF
predictDomains(new_query_gtf, Mmusculus, ncores=1)
#> Checking CDSs and translating protein sequences
#> Predicting domain families for 4 proteins
#> # A tibble: 14 × 5
#> transcript description eval begin end
#> <chr> <chr> <chr> <dbl> <dbl>
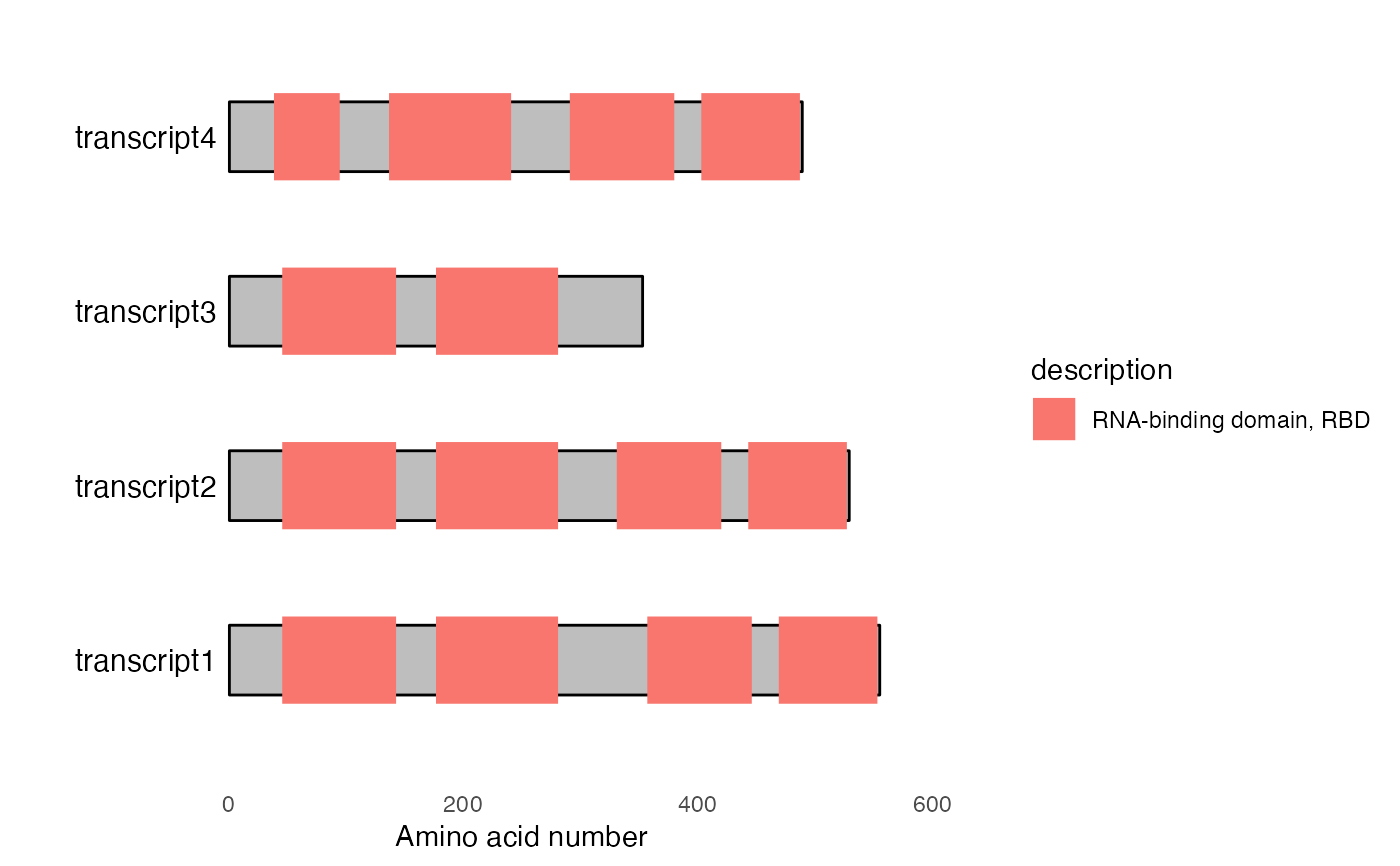
#> 1 transcript1 RNA-binding domain, RBD 1.54e-20 46 143
#> 2 transcript1 RNA-binding domain, RBD 1.54e-20 357 446
#> 3 transcript1 RNA-binding domain, RBD 1.54e-20 177 281
#> 4 transcript1 RNA-binding domain, RBD 1.54e-20 469 553
#> 5 transcript2 RNA-binding domain, RBD 1.54e-20 46 143
#> 6 transcript2 RNA-binding domain, RBD 1.54e-20 331 420
#> 7 transcript2 RNA-binding domain, RBD 1.54e-20 177 281
#> 8 transcript2 RNA-binding domain, RBD 1.54e-20 443 527
#> 9 transcript3 RNA-binding domain, RBD 1.54e-20 46 143
#> 10 transcript3 RNA-binding domain, RBD 1.54e-20 177 281
#> 11 transcript4 RNA-binding domain, RBD 1.54e-20 291 380
#> 12 transcript4 RNA-binding domain, RBD 1.54e-20 137 241
#> 13 transcript4 RNA-binding domain, RBD 1.54e-20 39 95
#> 14 transcript4 RNA-binding domain, RBD 1.54e-20 403 487
# predict domains of CDSs from Ptbp1 gene
predictDomains(new_query_gtf, Mmusculus, gene_name == "Ptbp1",ncores=1)
#> Checking CDSs and translating protein sequences
#> Predicting domain families for 4 proteins
#> # A tibble: 14 × 5
#> transcript description eval begin end
#> <chr> <chr> <chr> <dbl> <dbl>
#> 1 transcript1 RNA-binding domain, RBD 1.54e-20 46 143
#> 2 transcript1 RNA-binding domain, RBD 1.54e-20 357 446
#> 3 transcript1 RNA-binding domain, RBD 1.54e-20 177 281
#> 4 transcript1 RNA-binding domain, RBD 1.54e-20 469 553
#> 5 transcript2 RNA-binding domain, RBD 1.54e-20 46 143
#> 6 transcript2 RNA-binding domain, RBD 1.54e-20 331 420
#> 7 transcript2 RNA-binding domain, RBD 1.54e-20 177 281
#> 8 transcript2 RNA-binding domain, RBD 1.54e-20 443 527
#> 9 transcript3 RNA-binding domain, RBD 1.54e-20 46 143
#> 10 transcript3 RNA-binding domain, RBD 1.54e-20 177 281
#> 11 transcript4 RNA-binding domain, RBD 1.54e-20 291 380
#> 12 transcript4 RNA-binding domain, RBD 1.54e-20 137 241
#> 13 transcript4 RNA-binding domain, RBD 1.54e-20 39 95
#> 14 transcript4 RNA-binding domain, RBD 1.54e-20 403 487
# predict domains of CDSs from Ptbp1 gene and plot architecture out
predictDomains(new_query_gtf, Mmusculus, gene_name == "Ptbp1", plot = TRUE,ncores=1)
#> Checking CDSs and translating protein sequences
#> Predicting domain families for 4 proteins
 #> # A tibble: 14 × 5
#> transcript description eval begin end
#> <chr> <chr> <chr> <dbl> <dbl>
#> 1 transcript1 RNA-binding domain, RBD 1.54e-20 46 143
#> 2 transcript1 RNA-binding domain, RBD 1.54e-20 357 446
#> 3 transcript1 RNA-binding domain, RBD 1.54e-20 177 281
#> 4 transcript1 RNA-binding domain, RBD 1.54e-20 469 553
#> 5 transcript2 RNA-binding domain, RBD 1.54e-20 46 143
#> 6 transcript2 RNA-binding domain, RBD 1.54e-20 331 420
#> 7 transcript2 RNA-binding domain, RBD 1.54e-20 177 281
#> 8 transcript2 RNA-binding domain, RBD 1.54e-20 443 527
#> 9 transcript3 RNA-binding domain, RBD 1.54e-20 46 143
#> 10 transcript3 RNA-binding domain, RBD 1.54e-20 177 281
#> 11 transcript4 RNA-binding domain, RBD 1.54e-20 291 380
#> 12 transcript4 RNA-binding domain, RBD 1.54e-20 137 241
#> 13 transcript4 RNA-binding domain, RBD 1.54e-20 39 95
#> 14 transcript4 RNA-binding domain, RBD 1.54e-20 403 487
#> # A tibble: 14 × 5
#> transcript description eval begin end
#> <chr> <chr> <chr> <dbl> <dbl>
#> 1 transcript1 RNA-binding domain, RBD 1.54e-20 46 143
#> 2 transcript1 RNA-binding domain, RBD 1.54e-20 357 446
#> 3 transcript1 RNA-binding domain, RBD 1.54e-20 177 281
#> 4 transcript1 RNA-binding domain, RBD 1.54e-20 469 553
#> 5 transcript2 RNA-binding domain, RBD 1.54e-20 46 143
#> 6 transcript2 RNA-binding domain, RBD 1.54e-20 331 420
#> 7 transcript2 RNA-binding domain, RBD 1.54e-20 177 281
#> 8 transcript2 RNA-binding domain, RBD 1.54e-20 443 527
#> 9 transcript3 RNA-binding domain, RBD 1.54e-20 46 143
#> 10 transcript3 RNA-binding domain, RBD 1.54e-20 177 281
#> 11 transcript4 RNA-binding domain, RBD 1.54e-20 291 380
#> 12 transcript4 RNA-binding domain, RBD 1.54e-20 137 241
#> 13 transcript4 RNA-binding domain, RBD 1.54e-20 39 95
#> 14 transcript4 RNA-binding domain, RBD 1.54e-20 403 487